APE
A CLI, Java API and RESTful API for the automated generation of computational pipelines (scientific workflows) from large collections of computational tools.
Web platform for workflow exploration and benchmarking in bioinformatics
| Badges | |
|---|---|
| Packages and Releases |   |
| Build Status | |
| Documentation Status | |
| DOI | |
| License |  |
The Workflomics web platform aims to address the challenge faced by life science researchers who work with increasingly large and complex datasets and struggle to create optimal workflows for their data analysis problems.
The platform facilitates a "Great Bake Off" of computational workflows in bioinformatics by integrating bioinformatics tools and metadata with technologies for automated workflow exploration and benchmarking. This enables a systematic and rigorous approach to the development of cutting-edge workflows, specifically in the field of proteomics, to increase scientific quality, robustness, reproducibility, FAIRness, and maintainability.
The platform currently focuses on the proteomics domain. We aim to extend the platform to additional domains, e.g., metabolomics, genomics.
http://145.38.190.48/
Note: The website does not currently use HTTPS.
The Workflomics web interface is part of a larger infrastructure that includes a Postgres database, a Postgrest API, a RESTful APE service, etc. The architecture is presented in the figure below:
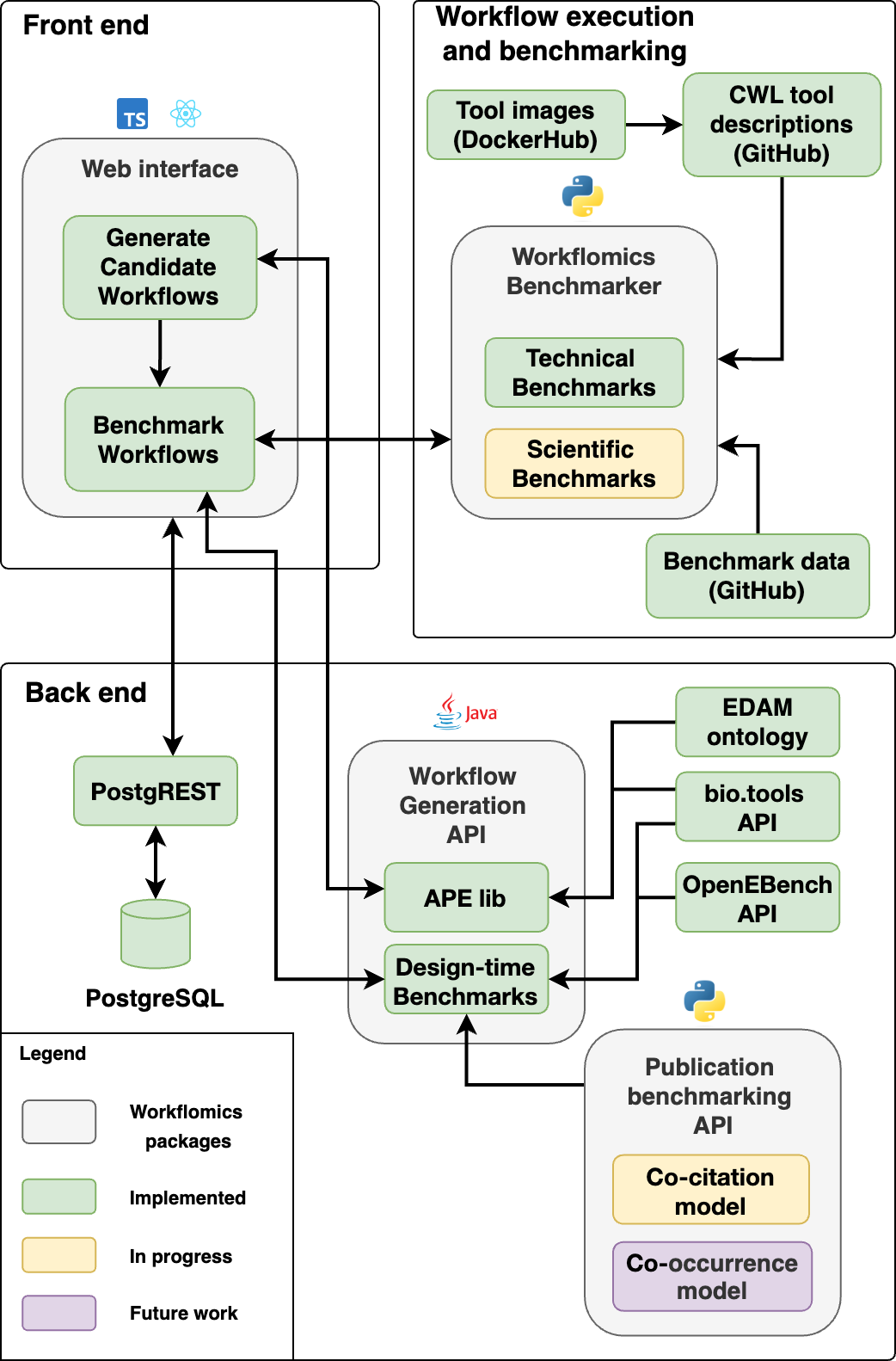

Workflomics: A platform for automated generation and workflow benchmarking in bioinformatics
A CLI, Java API and RESTful API for the automated generation of computational pipelines (scientific workflows) from large collections of computational tools.
RESTful API for the APE (Automated Pipeline Explorer) library.