Life Sciences

KNIME Pharmacophore
A plugin for the KNIME workflow system that adds a Pharmacophore data type and nodes to read, write, and align them.
- Workflow technologies
- (Java)

KNIME PLANTS
A node for the KNIME workflow system that allows you to use the PLANTS protein-ligand docking algorithm.
- Workflow technologies
- (Java)
- (Mustache)

KNIME Python node archetype
Want to write your own KNIME node wrapping a Python library. Then use the KNIME Python node archetype to generate a node skeleton repository with sample code
- Big data
- Workflow technologies
- (Java)
- (Python)

KNIME Python wrapper
If you are developing a KNIME node which calls some Python code, then the KNIME Python wrapper can help you.
- Workflow technologies
- (Java)
- (Python)

KNIME Silicos-it nodes
A node for the KNIME workflow systems that allows you to use the Silicos-it software to filter or align molecules.
- Workflow technologies
- (Java)

3D-e-Chem Virtual Machine
A freely available Virtual Machine encompassing tools, databases, and workflows for cheminformaticians, including new resources developed for ligand binding site comparisons and GPCR research.
- Inter-operability & linked data
- Workflow technologies
- (Shell)
- (Smarty)
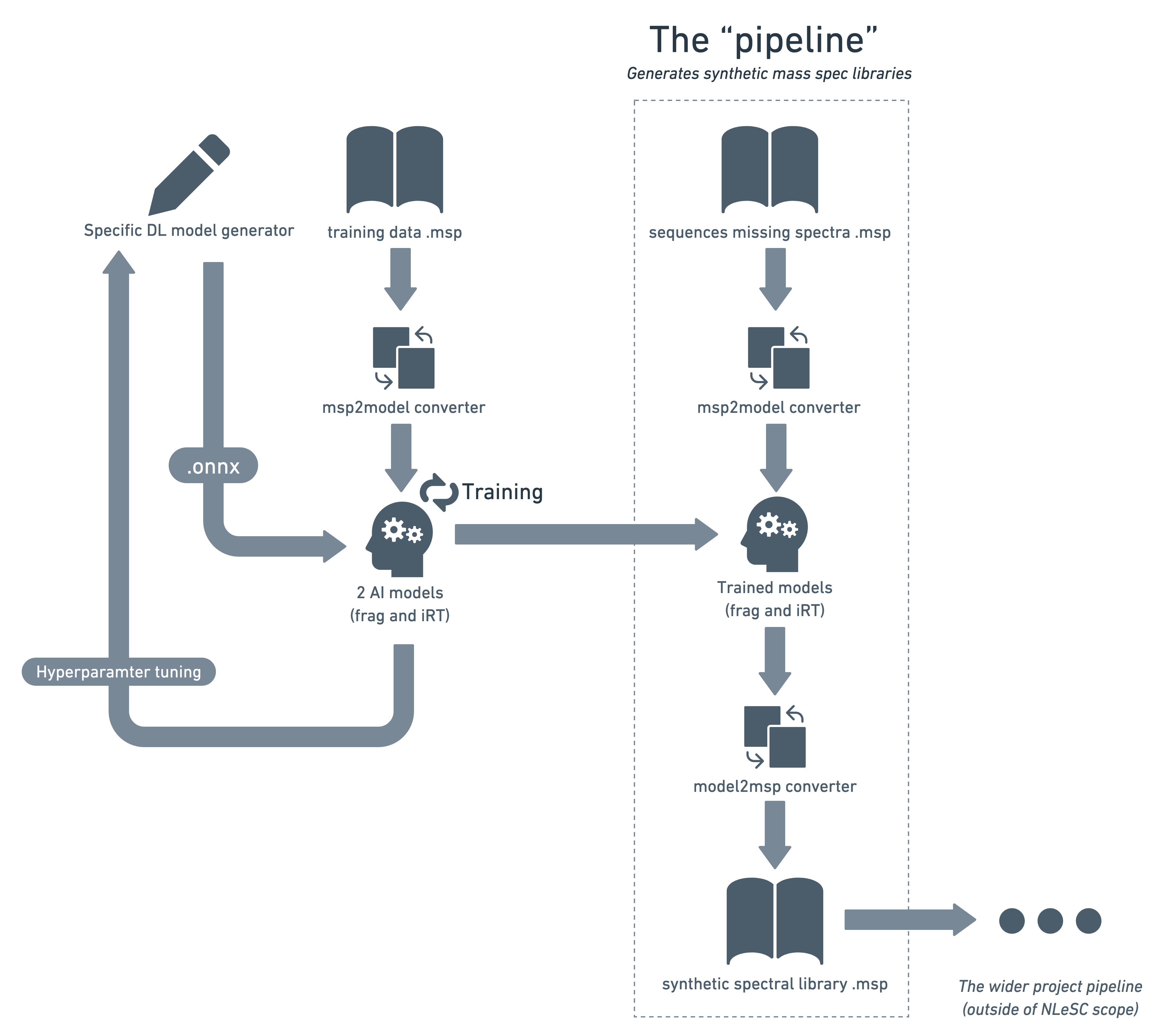
aiproteomics
Generate and compare deep learning models for generating synthetic mass spectral libraries
- Machine learning
- mass spectrometry
- phosphoproteomics
- + 1
- ("Jupyter Notebook")
- (Python)
- (Shell)

BestieTemplate.jl
BestieTemplate.jl is a template focused on best practices for package development in Julia.
- best practices
- julia
- template
- (Julia)
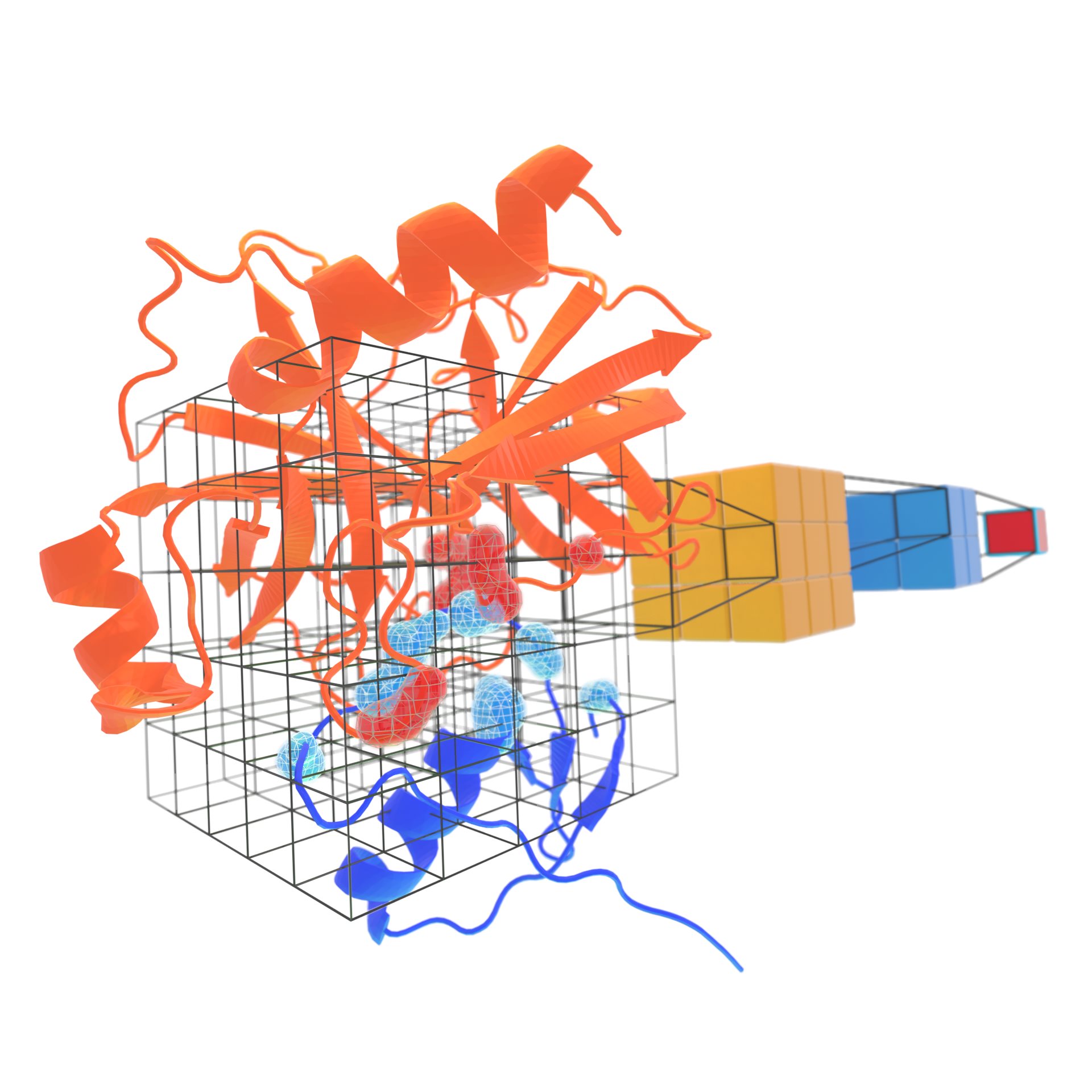
DeepRank
Deep learning framework for data mining protein-protein interactions using CNN
- Big data
- Machine learning
- Optimized data handling
- (C)
- (Makefile)
- (Python)
- + 2
DeepRank2
DeepRank2 is an open-source deep learning framework for data mining of protein-protein interfaces or single-residue missense variants. This package is an improved and unified version of three previously developed packages: DeepRank, DeepRank-GNN and DeepRank-Mut.
- Cancer vaccine
- Convolutional Neural Network
- Deep Learning Framework
- + 5
- (Dockerfile)
- ("Jupyter Notebook")
- (Python)
- + 1
DeepRank GNN
DeepRank-GNN is the graph neural network of our DeepRank package. DeepRank GNN allows to train graph neural networks to classify protein-protein interface
- Machine learning
- Protein-Protein Interaction
- (Python)
Dive
Interactively explore millions of 2D and 3D data points in your browser, without the need to install anything.
- Big data
- Machine learning
- Visualization
- (CSS)
- (HTML)
- (JavaScript)
- + 1