matchms
MA
Python library for fuzzy comparison of mass spectrum data and other Python objects
Updated 29 months ago
134 14
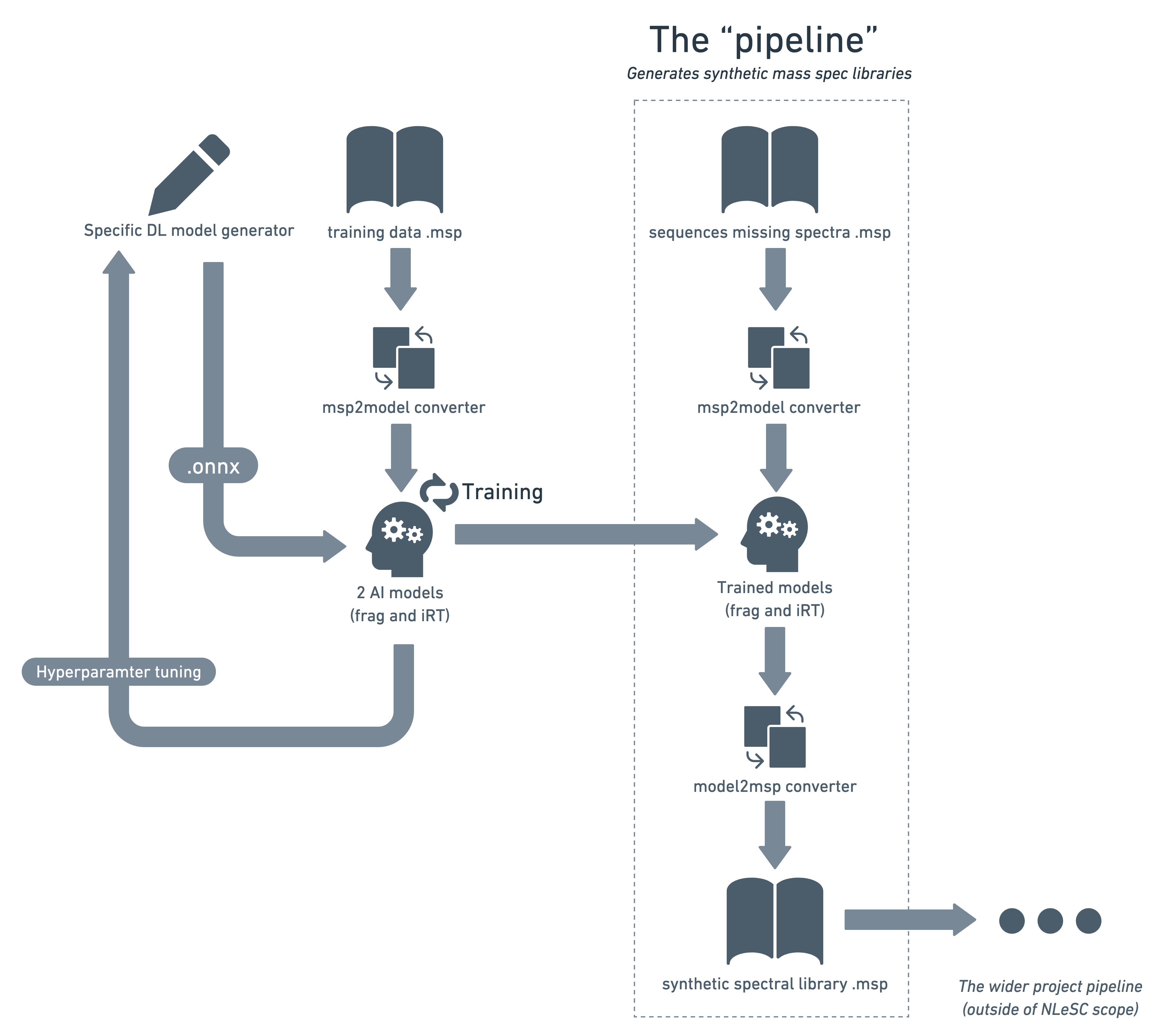
Generate and compare deep learning models for generating synthetic mass spectral libraries
This package contains various tools, datasets and ML model implementations from the field of (phospho-)proteomics. It is intended to facilitate the testing and comparison of different neural network architectures and existing models, using the same datasets. Both retention time and fragmentation (MSMS) models are included.
Implementations of existing models from the literature are intended to be modifiable/extendable. For example, so that tests may be carried out with different peptide input lengths etc.
Python library for fuzzy comparison of mass spectrum data and other Python objects