All software

compareMS2
compareMS2 is a tool for direct comparison of tandem mass spectrometry datasets, typically from liquid chromatography-tandem mass spectrometry (LC-MS/MS), defining a distance as a function of the share of similar spectra. Applications span biomedicine, phylogenetics, forensics and food science.
- Forensics
- mass spectrometry
- phylogenetics
- + 1
- (C)
- (CSS)
- (HTML)
- + 2
PIQMIe
A web tool for mass spectrometry-based proteomics data analyses
- Data Analysis
- mass spectrometry
- Optimized data handling
- + 2
- (Dockerfile)
- (HTML)
- (JavaScript)
- + 3
mzRecal
mzRecal recalibrates mass spectrometry (MS1) data in mzML format, using peptide identifications in mzIdentML.
- mass spectrometry
- proteomics
- (Dockerfile)
- (Go)
- (Makefile)
- + 2

OpenMS
OpenMS is an open-source software C++ library for LC-MS data management and analyses. It offers an infrastructure for rapid development of mass spectrometry related software. OpenMS is free software available under the three clause BSD license and runs under Windows, macOS, and Linux.
- C++
- mass spectrometry
- metabolomics
- + 1
- (Batchfile)
- (C)
- (C++)
- + 15
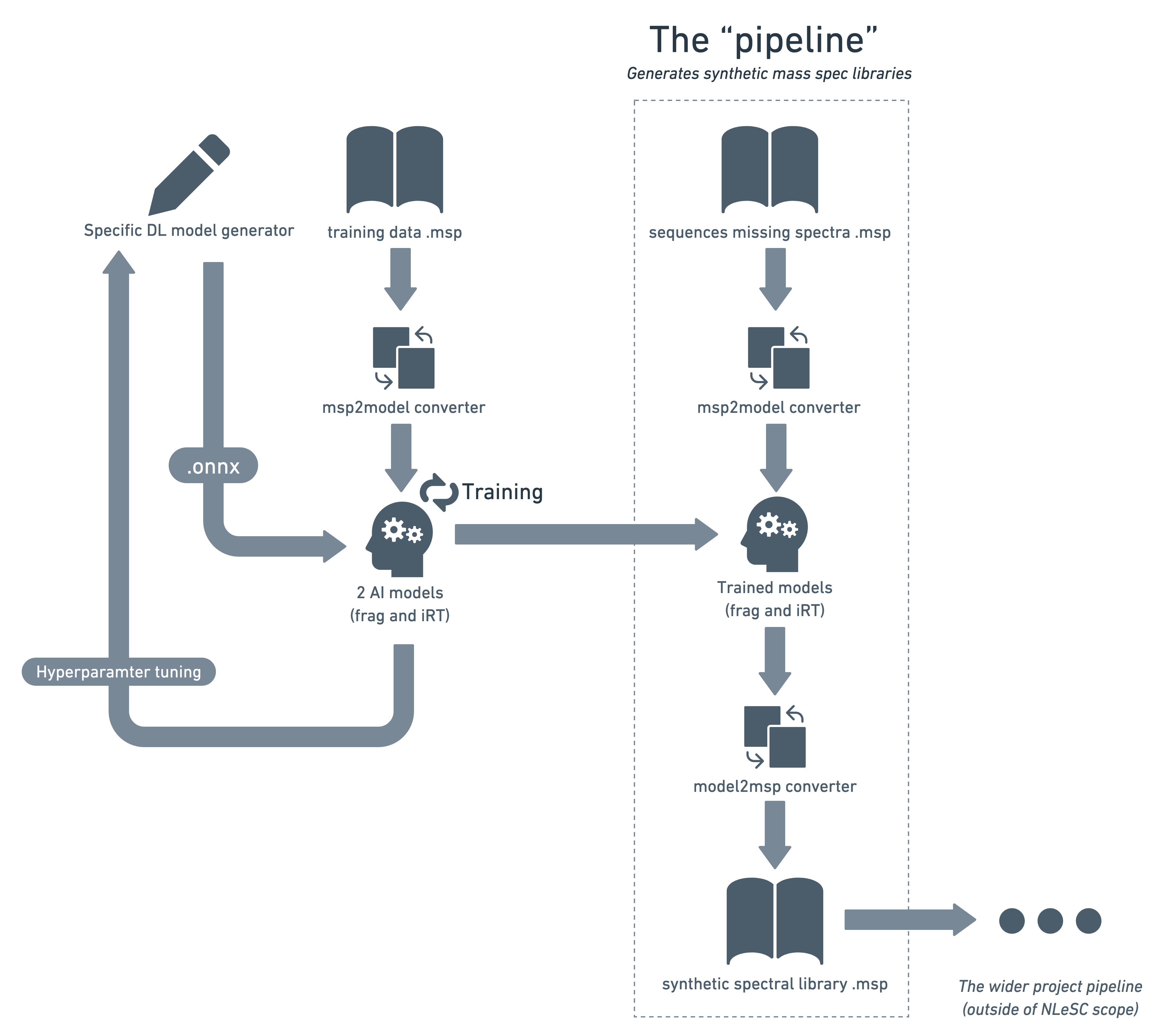
aiproteomics
Generate and compare deep learning models for generating synthetic mass spectral libraries
- Machine learning
- mass spectrometry
- phosphoproteomics
- + 1
- ("Jupyter Notebook")
- (Python)
- (Shell)

Mass Spec Studio for Clinical Proteomics
A fusion of the popular OpenMS suite of functionality with the powerful Mass Spec Studio. Users have access to OpenDIA and OpenPRM to support large scale processing of clinical proteomics data sets.
- mass spectrometry
- proteins
- proteomics