Stefan Verhoeven

haddock3
Haddock3 (High Ambiguity Driven protein-protein DOCKing) is a modular software for for the integrative modeling and refinement of biomolecular complexes.
- biomolecular complexes
- docking
- energetics
- + 4
- (BitBake)
- (C)
- (C++)
- + 11

ESMValTool
The Earth System Model eValuation Tool is a community diagnostics and performance metrics tool for the evaluation of Earth System Models that allows for routine comparison of models and observations.
- Big data
- Optimized data handling
- Visualization
- + 1
- (Dockerfile)
- ("Emacs Lisp")
- (Jinja)
- + 7

spec2vec
spec2vec is a novel similarity measure for comparing mass spectrometry data, which learns peak representations using Word2Vec.
- Machine learning
- Text analysis & natural language processing
- (Batchfile)
- (Makefile)
- (Python)

matchms
Python library for fuzzy comparison of mass spectrum data and other Python objects
- Big data
- Optimized data handling
- (Batchfile)
- (Makefile)
- (Python)
- + 1
Paired omics data platform
If you do metabolomics experiments with mass spectra and have sequenced the genomes of the samples, then the platform can help you link them.
- Inter-operability & linked data
- (CSS)
- (Dockerfile)
- (HTML)
- + 3
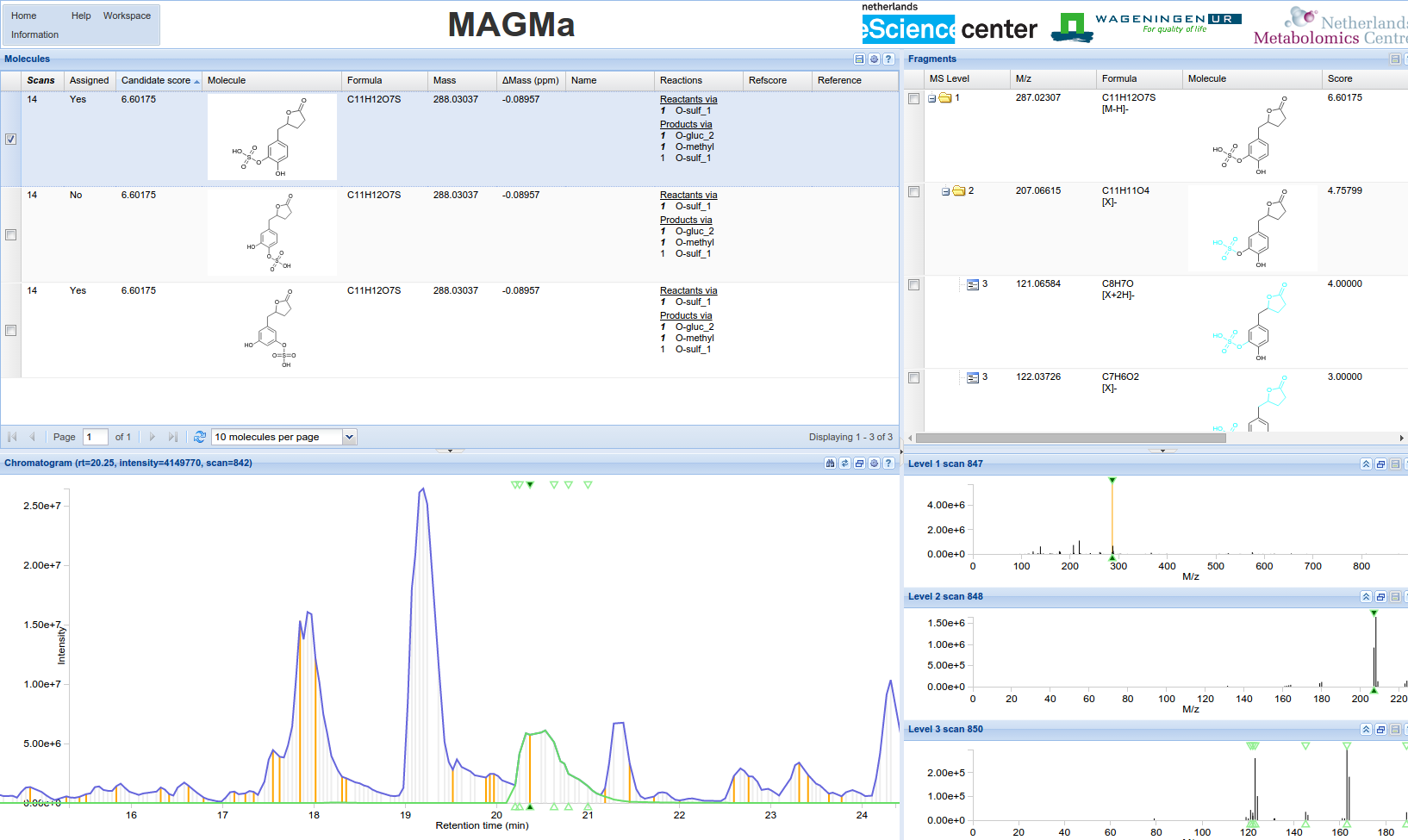
MAGMa
MAGMa is an online application for the automatic chemical annotation of mass spectrometry data.
- Big data
- Visualization
- (CSS)
- (Cython)
- (Dockerfile)
- + 5

powerfit
Rigid body fitting of atomic strucures in cryo-electron microscopy density maps
- cryo-electron microscopy
- macromolecular structure
- map fitting
- (C)
- (Cython)
- (Dockerfile)
- + 1

Netherlands eScience Center Python Template
Generic template for Python packages, so you can spend less time setting up and configuring, and comply with the Netherlands eScience Center Software Development Guide from the start.
- software development tools
- software sustainability
- template
- (Jinja)
- (Python)
- (Shell)
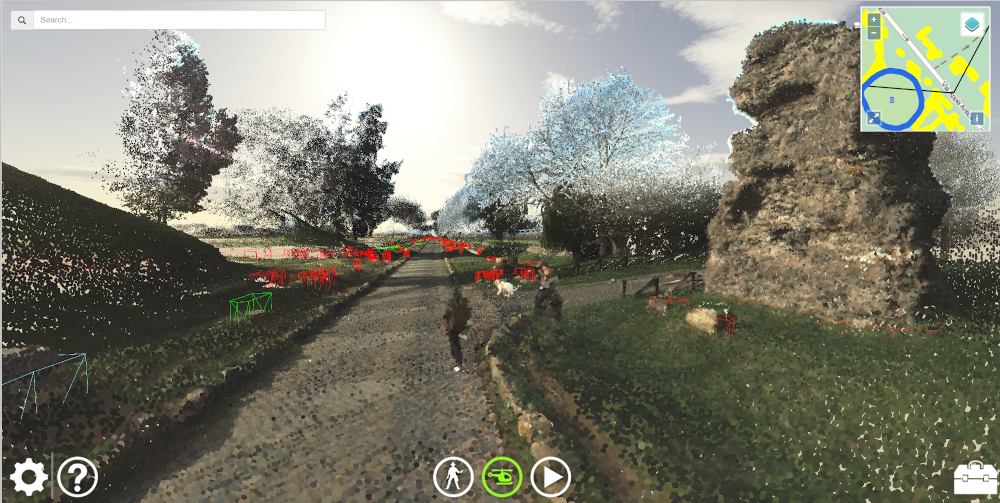
Via Appia Visualization
A web application which visualizes the point clouds and additional data gathered in the Via Appia project with WebGL.
- Visualization
- (CSS)
- (HTML)
- (JavaScript)
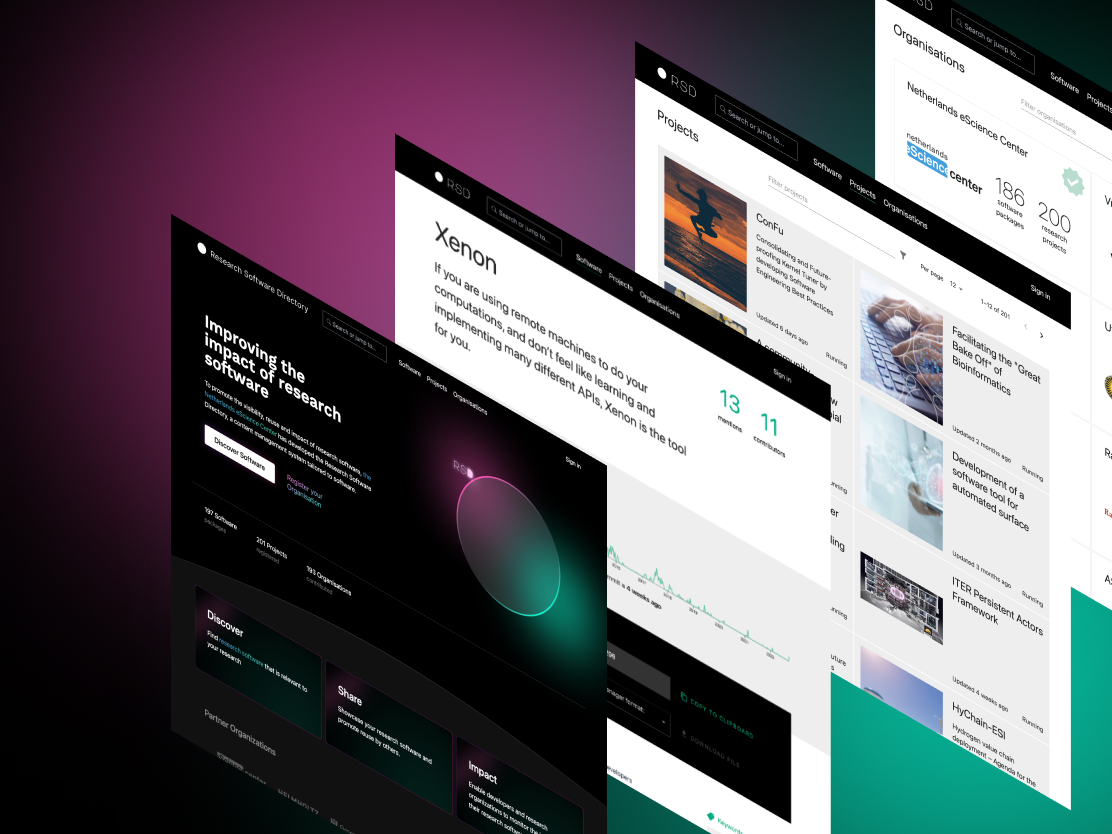
Research Software Directory
A content management system for research software, which promotes the visibility, reuse, and impact of research software.
- FAIR
- Research Software Impact
- Software Citation
- (CSS)
- (Dockerfile)
- (Go)
- + 9

3D-e-Chem Virtual Machine
A freely available Virtual Machine encompassing tools, databases, and workflows for cheminformaticians, including new resources developed for ligand binding site comparisons and GPCR research.
- Inter-operability & linked data
- Workflow technologies
- (Shell)
- (Smarty)

KLIFS KNIME nodes
If you are working in the KNIME worflow platform and need data about your favorite kinase receptor ligand interaction, then these nodes are for you.
- Workflow technologies
- (Java)
- (Scala)
- (Shell)